A companion video has been to our paper, “Prediction of Mutant mRNA Splice Isoforms by Information Theory-Based Exon Definition,”
DOI: 10.1002/humu.22277, has been published on the Human Mutation website.
A companion video has been to our paper, “Prediction of Mutant mRNA Splice Isoforms by Information Theory-Based Exon Definition,”
DOI: 10.1002/humu.22277, has been published on the Human Mutation website.
Experience the superior sensitivity and speed of the pipeline for yourself. After downloading the free client, you can connect to a trial version of Cytognomix’s commercial plugin installed on the CLC Bio Genomics Server on our DAIR cloud account. There, you may work with the results of our genome-wide analysis of three different cancer cell lines (described in our upcoming paper, see below), or import your own variants and analyze them. Download the Trial Shannon Pipeline Installation Guide for further details on the capabilities of the trial version and on how you may access it.
“Intensity Integrated Laplacian Based Thickness Measurement for Detecting Human Metaphase Chromosome Centromere Location” by Akila Subasinghe Arachchige, Jagath Samarabandu, Joan Knoll and Peter Rogan will be published in IEEE Transactions in Biomedical Engineering.
Abstract—Accurate detection of the human metaphase chromosome centromere is an important step in many chromosome analysis and medical diagnosis algorithms. The centromere location can be utilized to derive information such as thechromosome type, polarity assignment etc. Methods available in literature yield unreliable results mainly due to high variabilityof morphology in metaphase chromosomes and boundary noise present in the image. In this article we have proposed a multi-staged algorithm which includes the use of discrete curve evolution (DCE), gradient vector flow (GVF) active contours, functional approximation of curve segments and support vector machine (SVM) classification. The standard Laplacian thickness measurement algorithm was enhanced to incorporate both contour information as well as intensity information to obtain a more accurate centromere location.
US Patent Application Serial No. ![]() 13/469,531 has had all claims allowed by the USPTO.
13/469,531 has had all claims allowed by the USPTO.
This application, AB INITIO GENERATION OF SINGLE COPY GENOMIC PROBES, covers the method and applications of single copy probes containing at least one divergent repetitive element. Examples of probes are described in our new publication: “Expanding probe repertoire and improving reproducibility in human genomic hybridization” by Stephanie N. Dorman, Ben C. Shirley, Joan H. M. Knoll and Peter K. Rogan in Nucleic Acids Research. The patent covers many uses of these probes, including FISH, microarray genomic hybridization, microsphere solution hybridization, and targeted capture arrays for genomic library enrichment in next generation sequencing.
“Automating Dicentric Chromosome Detection from Cytogenetic Biodosimetry Data” at the International EPRBioDose 2013 Conference in Leiden, Netherlands (March 24-28).
Authors: Peter Rogan(1,2), Akila Subasinghe(1), Asanka Wickramasinghe(1), Yanxin Li(1), Jagath Samarabandu(1), Joan Knoll(1,2), Ruth Wilkins(3), Farah Flegal(4); (1)University of Western Ontario, (2)Cytognomix Inc., (3)Health Canada, (4)Atomic Energy of Canada Ltd., Canada.
Abstract: We are developing a prototype software system with sufficient capacity and speed to estimate radiation exposures by counting dicentric chromosomes in metaphase cells from many individuals in the event of a
mass casualty. Top-ranked metaphase images are segmented by defining chromosomes with an active contour gradient vector field (GVF), and by determining centromere locations along the centerline. The centerline is
extracted by Discrete Curve Evolution (DCE) skeleton branch pruning and curve interpolation. Centromere detection minimizes the global width and DAPI-staining intensity profiles along the centerline. A second
centromere is identified by reapplying this procedure after masking the first. Dicentrics can be identified by applying a support vector machine-based classification, which uses features that capture width and intensity
profile characteristics as well as local shape features of the object contour at candidate pixel locations. The correct location of the centromere is also refined in chromosomes with sister chromatid separation. The
overall algorithm has both high sensitivity (85%) and specificity (94%). Results are independent of the shape and structure of chromosomes in different cells, regardless of which laboratory protocol is followed or the
specimen source. The requisite throughput is being achieved by recoding MATLAB software modules for different segmentation functions in C++/OpenCV, and integrating them in the prototype. Processing of
numerous images is accelerated by both data and task software parallelization with the Message Passaging Interface and Intel Threading Building Blocks as well as an asynchronous non-blocking I/O strategy. Relative
to a serial process, metaphase ranking, GVF, and DCE are respectively 100 and 300 fold faster on an 8-core I7-based desktop and on a 64-core shared memory cluster computer. Extrapolation from these benchmarks to
a 64-core system in which all of the software modules have been integrated indicates that it should be feasible to process metaphases for dicentric chromosomes from 1000 specimens in 20 hours.
Interpretation, stratification and validation of sequence variants affecting mRNA splicing in complete human genome sequences. Genomics, Proteomics, and Bioinformatics, 11:77-85, 2013.
Ben C. Shirley, Eliseos J. Mucaki, Tyson Whitehead, Paul I. Costea, Pelin Akan, Peter K. Rogan.
Information theory-based methods have been shown to be sensitive and specific for predicting and quantifying the effects of non-coding mutations in Mendelian diseases. We present and validate the Shannon pipeline software to predict mRNA splicing mutations for genome-scale analysis. Individual information contents (in bits) of reference and variant splice sites are compared and significant differences are annotated and prioritized. The software has been implemented for CLC-Bio Genomics platform as the Shannon pipeline. Annotation indicates the context of novel mutations as well as common and rare SNPs with splicing effects. Potential natural and cryptic mRNA splicing are identified, and null mutations are distinguished from leaky mutations. Mutations and rare SNPs were predicted in 3 cancer cell line genomes (U2OS, U251, and A431), and then experimentally validated by expression analyses. After filtering, tractable numbers of potentially deleterious variants are predicted by the software, suitable for further laboratory investigation. In these cell lines, novel functional variants comprised between 6 to 17 inactivating and 1 to 5 leaky mutations, and 6 to 13 cryptic splicing mutations. Predicted effects were confirmed by RNAseq analysis of U2OS, U251, and A431 cell lines, and expression microarray analysis of SNPs in HapMap cell lines.
“Relating centromeric topography in fixed human chromosomes to alpha-satellite DNA and CENP-B distribution:
Authors: Khan W., Chisholm R., Tadayyon S., Subasinghe A., Norton P., Samarabandu J., Johnston LJ., Knoll JH., Rogan PK.
“Expanding probe repertoire and improving reproducibility in human genomic hybridization” by S. Dorman, B. Shirley, J. Knoll, and P. Rogan has been accepted for publication by the journal Nucleic Acids Research. In this paper, we use Cytognomix’s patented ab initio sc probe technology to develop and validate a novel classes of FISH probes, genomic microarrays and oligonucleotide pools useful for targeted capture hybridization.
High-throughput sequencing (HTS) of both healthy and disease singletons yields many novel and low frequency variants of uncertain significance (VUS). Some non-coding sequence variants have been proven to significantly contribute to the phenotypes of high penetrance disorders. We develop an approach to predict pathogenicity of non-coding VUS based on comprehensive information analysis of changes in DNA and RNA sequences bound by regulatory factors. Using cleavable solution microarrays, we are capturing and enriching for non-coding variants in genes known to harbor mutations that increase breast cancer risk. Oligo baits covering ATM, BRCA1, BRCA2, CDH1, CHEK2, PALB2 and TP53 were synthesized for solution hybridization with a custom cleavable microarray spanning the complete coding and intergenic regions 10 kb upstream and downstream of each gene. Non-exonic sequences are densely populated with repetitive sequences that can affect short read assembly. A novel probe design method was used to capture both repeat-free and divergent repeat sequences that are effectively single copy. After SBS sequencing of 13 patient samples in our laboratory, information theory-based sequence analysis was used to prioritize non-coding variants which occurred within sequence elements recognized by proteins or protein complexes. The novel VUS identified are being investigated for effects on mRNA splicing, transcription factor-binding site (TFBS), and untranslated region (UTR) mutations. We have developed and apply information theory based models for exon recognition, which predict the relative abundance of natural, cryptic, and mutant splice isoforms resulting from predicted mutations using the combined donor and acceptor site strengths of each mRNA species. We have applied a similar approach to detect mutations in the promoters of BRCA1 and BRCA2 that alter strengths of TFBS. Information weight matrices were automatically computed by entropy minimization of ATF3, BATF, BCL3, BCLAF, c-Jun, c-Myc, CTCF, EGR1, EP300, ETS1, FOSL2, FOXA1, FOXM1, GABP, GATA3, GRP20, HSF1, IRF4, MEF2A, NFIC, NFkB, PU.1, RAD21, RXRA, TCF12, TCF7L2, and YY1 TFBS from the global set of ENCODE ChiP-seq regions embedded within DNAse I hypersensitive domains. These models were then used to evaluate novel variants discovered by sequence analysis of breast cancer patients for alteration the TFBS binding strengths. This strategy more comprehensively covers non-coding regions in breast cancer genes than repeat masking, and introduces a unified framework for systematic interpretation of VUS that may affect expression.
Cytognomix has established partnerships to develop advanced high throughput software and microscope systems to automate cytogenetic biodosimetry in the event of a mass casualty radiation event. Biodosimetry laboratory partners include Health Canada and Atomic Energy of Canada Ltd. Software development for image processing of chromosome images is being carried out at the Schools of Engineering and Schulich School of Medicine and Dentistry, Western University, Ontario CA. We are providing software to Huron Technologies International, Waterloo CA to accelerate the detection of dicentric metaphase chromosomes using a modification of their MACROscope system. The project has been supported by a Western Innovation Grant, a CMCR pilot project (NIH subcontract from Dartmouth University), and by the Federal Development Agency of Southern Ontario.
Read this description of our project with Huron Technologies: ApplicationNote_cytogenetic_biodosimetry
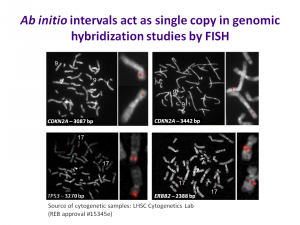 US Patent 8,209,129 has issued on single copy DNA probes which include divergent repetitive sequences, thus significantly extending the portions of the genome that can be used for such probes beyond traditional single copy sequences.
US Patent 8,209,129 has issued on single copy DNA probes which include divergent repetitive sequences, thus significantly extending the portions of the genome that can be used for such probes beyond traditional single copy sequences.
The technology also increases the density of genomic DNA probes for higher resolution genetic analysis beyond what is used in FISH, genomic microarrays for array comparative genomic hybridization, and solution capture hybrdization arrays for sequence enrichment in deep sequencing. It is licensed to Cytognomix.
Cytognomix receives US Patent 7,734,424 entitled “AB INITIO GENERATION OF SINGLE COPY GENOMIC PROBES.”
Ontario Centre of Excellence for Commercialization of Research assists Cytognomix in building capacity through its Embedded Executive Program.
/
Canadian Institute for Photonic Innovations awards grant to the University of Western Ontario and Cytognomix through their Technology Exploitation and Networking Program.
MaRS Ventures Inc. (Toronto) accepts Cytognomix as a client of the Business Mentorship Entrepreneurship Program.
Cytognomix becomes a client of the NRC-IRAP program and receives an initial investment to create jobs in Ontario.