Tag Archives: interpretation of SNPs
Feb. 11, 2020. Article about allele-specific alternative splicing published
Mucaki, E.J., Shirley, B.C. and Rogan, P.K., 2020. Expression changes confirm genomic variants predicted to result in allele-specific, alternative mRNA splicing. Frontiers in Genetics, 11: 109 (https://www.frontiersin.org/articles/10.3389/fgene.2020.00109/full).
(preprint in bioRxiv, 549089 [doi: https://doi.org/10.1101/549089])
Sept. 7, 2019. Pan cancer article and database update
March 20, 2019. Presentation at the 2019 American College of Medical Genetics and Genomics annual conference
The following paper has been accepted for presentation:
“Pan-Cancer Repository of Validated Natural and Cryptic mRNA Splicing Mutations”,
Category: “Laboratory genetics and genomics”, Abstract Poster Number: 754 (link to Abstract)
Where: Exhibit hall, Washington Convention Center, ACMG Clinical Genetics Meeting in Seattle, Washington
When: April 2 – 6, 2019; Poster presentation time: Friday, 4/5 from 10:30am-12:00pm
This work will be available as an ePoster AS WELL AS being presented in printed format on a poster board during the Annual Meeting. Details to access the ePoster will be available soon.
March 17, 2015. Review article on mRNA splicing mutations highlighted on F1000Research blog
Revised version of our review article on mRNA splicing in F1000Research has been highlighted in the Faculty of 1000 blog:
http://blog.f1000research.com/2015/03/17/interpreting-genomic-variants-in-rare-and-common-diseases/
The blog entry includes links to our YouTube video:
Short version (3:42)
Long version (9:41)
Mutation Forecaster®
MutationForecaster® (mutationforecaster.com) is Cytognomix’s patented web-portal for analysis of all types of mutations (coding and non-coding), including interpretation, comparison and management of genetic variant data. It’s a fully automated genome interpretation solution for research, translational and clinical labs.
MutationForecaster® combines our world-leading genome interpretation software on your exome, gene panel, or complete genome (Shannon transcription factor and splicing pipelines, ASSEDA, Veridical) with the Cytognomix User Variation Database and Variant Effect Predictor. With our integrated suite of software products, analyze coding, non-coding, and copy number variants, and compare new results with existing or your own database. Select predicted mutations by phenotype using articles with Cyto Visualization Analytics. With Workflows, automatically perform end-to-end analysis with all of our software products.
Download an 1 page overview of MutationForecaster®: link .
You can now experience our integrated suite of genome interpretation products through a free trial of MutationForecaster®. Once you register, analyze datasets that we have analyzed in our peer-reviewed publications with any of our software tools.
To obtain a subscription (2 months or 1 year [pricing]) to MutationForecaster®, please contact us.
Don’t want to run your own analyses? Let us do it for you with our Bespoke Analysis Service.
Resources
Links to the latest CytoGnomix products:
Applications and consulting in Geostatistical Epidemiology
Monitoring and discriminating infectious disease hotspots from high disease burden regions, eg. for COVID-19:
Zenodo repository: Geostatistical Analysis of SARS-CoV-2 Positive Cases in the United States
Defence Canada IDEaS project: Locating emerging COVID19 hotspots in Ontario after community transmission by time-correlated, geospatial analysis
Addressing large scale radiation incidents and accidents:
Article in PLOS One: Meeting radiation dosimetry capacity requirements of population-scale exposures …. (Funded by High performance computing consortium: SOSCIP and CytoGnomix)
How to: Protocol for Geostatistical Determination of Radiation Dosimetry Maps of Population-Scale Exposures
Large scale Radiation Biodosimetry
Capacity of supercomputer version of Automated Dicentric Chromosome Identifier and Dose Estimator (ADCI) software: Automated Cytogenetic Biodosimetry at Population-Scale and Radiation, Radiation, 2021 (link to published article).
Scalable, democratized access to ADCI:
Overview of Cloud version- ADCI_Online
Presentation to the International Atomic Energy Agency (CRP E35010)
Gene Expression Signatures for Radiation Biodosimetry
Mucaki, E.J., Shirley, B.C. and Rogan, P.K., 2021. Improved radiation expression profiling in blood by sequential application of sensitive and specific gene signatures. International Journal of Radiation Biology, doi.org/10.1080/09553002.2021.1998709 Link to pdf: Improved radiation expression profiling…
Zhao, J.Z., Mucaki, E.J. and Rogan, P.K., 2018. Predicting ionizing radiation exposure using biochemically-inspired genomic machine learning. F1000Research, 7(233), p.233. Link to open access article: https://f1000research.com/articles/7-233
Large Scale Repository of Cancer Splicing Mutations
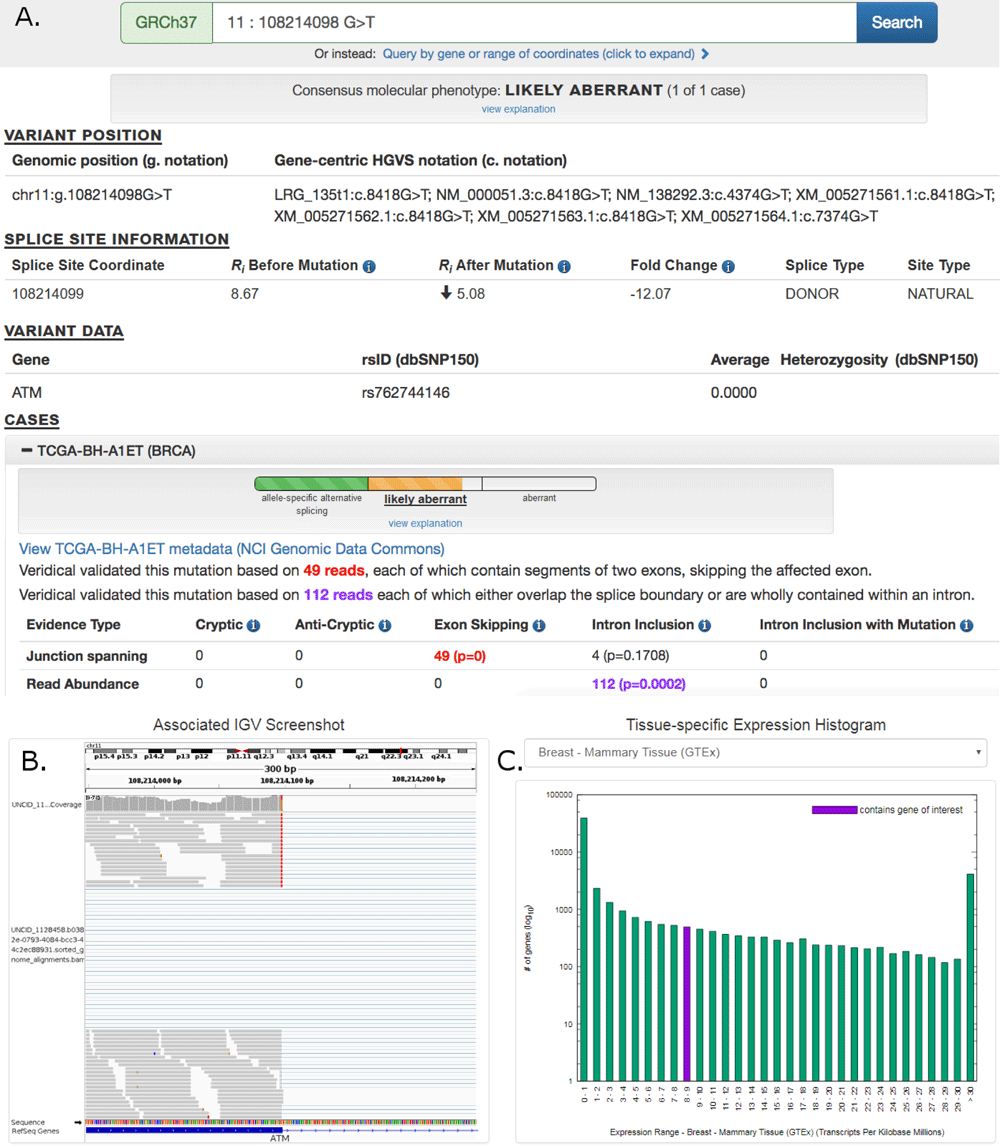
Pan-cancer repository of validated natural and cryptic mRNA splicing mutations (a major public resource of mRNA splicing mutations validated according to multiple lines of evidence of abnormal gene expression. )
Article in F1000Research: Pan-Cancer repository of …..
Presentation at the 2019 American College of Medical Genetics Annual Meeting:
Pan-cancer repository of validated natural and cryptic mRNA.ePoster
Interactive Website: Gene signatures for chemotherapy drug response
Demo (Windows): Automated Dicentric Chromosome Identifier and Dose Estimator
Review on information theory-based splicing mutation analysis:
Caminsky et al. 2014, Videos describing this paper: short and long versions.