On May 10, 2021, CytoGnomix is presenting a poster at ConRad 2021 (www.radiation-medicine.de) titled:
Demonstration of the Automated Dicentric Chromosome Identifier and Dose Estimator [ADCI] System in a Cloud-based, Online Environment.
From the abstract:
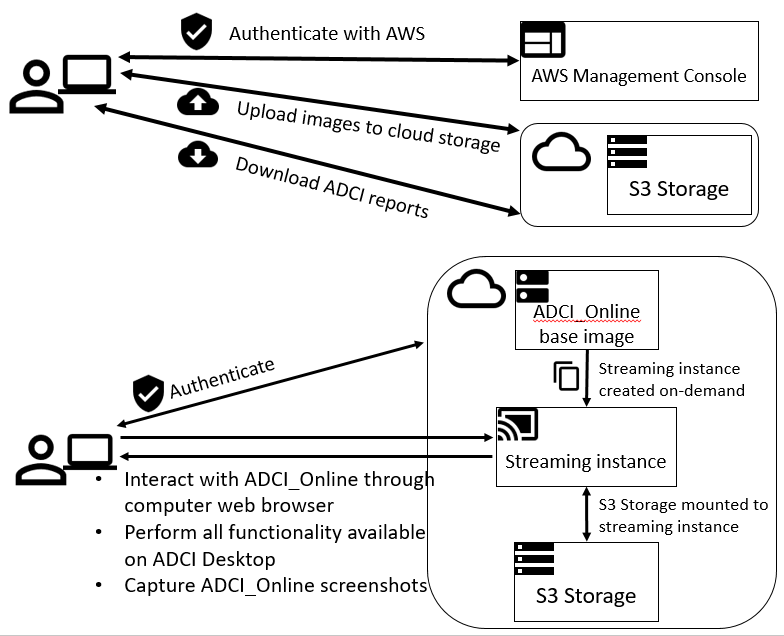
Interpretation of cytogenetic metaphase images and quantification of exposures remain labour intensive in radiation biodosimetry, despite computer-assisted dicentric chromosome (DC) recognition and strategies to share workloads among different biodosimetry laboratories. ADCI processes the captured images to identify DCs, selects images, and quantifies radiation exposure. This paper describes ADCI_Online, a secure web-streaming platform on Amazon Web Services that can be accessed worldwide from distributed local nodes.
ADCI_Online offers a subscription-based service useful for radiation research, biodosimetry proficiency testing, inter-laboratory comparisons, and training. In a research context, the system could provide highly uniform, reproducible assessment in large studies of many individuals, for example, exposed to therapeutic radiation. ADCI_Online compute environments originate from a single snapshot which can be cloned any number of times; thus, the system can be rapidly scaled when required. With robust network connectivity in a medical emergency of multiple potentially radiation exposed individuals, throughput and capacity for multiple samples requiring simultaneous processing and dose evaluation can be expanded to seamlessly mitigate any backlog in sample interpretation.