Hill, S.L., Rogan, P.K., Wang, Y.X. Knoll J.H.M. Differentially accessible, single copy sequences form contiguous domains along metaphase chromosomes that are conserved among multiple tissues. Mol Cytogenet 14, 49 (2021). https://molecularcytogenetics.biomedcentral.com/articles/10.1186/s13039-021-00567-w
Tag Archives: metaphase chromosomes
July 4, 2019. Presentations describing interlaboratory comparison of radiation exposure determination by automated cytogenetic biodosimetry
We will be presenting:
Determination of radiation exposure levels by fully automated
dicentric chromosome analysis: Results from IAEA MEDBIODOSE
(CRP E35010) interlaboratory comparison
at both the 19th International Congress of Radiation Research (Aug. 25-29, 2019) and the 12th International Symposium on Chromosome Aberrations (Aug. 27, 2019) in Manchester, UK. This study compared the performance of our Automated Dicentric Chromosome Identifier and Dose Estimator (ADCI) using data from 6 different laboratories. Each of these members of the IAEA-sponsored Cooperative Research Project E35010, submitted images for calibration curve construction and at least 2 samples of unknown exposure to CytoGnomix for analysis with ADCI. We will report the results of this analysis during this presentation.
This poster presentation is now available on the Zenodo website (http://doi.org/10.5281/zenodo.4012749)
doi: DOI 10.5281/zenodo.4012748
Authors:
Rogan P , Shirley B , Li Y , Guogyte K , Sevriukova O , Ngoc Duy P , Moquet J ,
Ainsbury E , Sudprasert W , Wilkins R , Norton F , Knoll J
Department of Biochemistry , University of Western Ontario, London Ontario, Canada
Department of Pathology and Laboratory Medicine, University of Western Ontario, London
Ontario, Canada
Radiation Protection Centre, Ministry of Health (L T -RPC), Vilnius, Lithuania
Dalat Nuclear Research Institute (VN-DNRI), Dalat, Vietnam
Public Health England (PHE), Oxford, Great Britain
Thai Biodosimetry Network, Kasetsart University (THA), Bangkok, Thailand
Health Canada, Ottawa Ontario, Canada
Canadian Nuclear Laboratories, Chalk River Ontario, Canada
Cytognomix, London Ontario, Canada
July 13, 2014. Presentations at the 2014 American Society of Human Genetics Conference
Cytognomix will be presenting several papers at the upcoming ASHG annual meeting (October 18-22, 2014, San Diego):
Using information theory to analyze and predict splicing mutations in rare and common diseases: performance and best practices. N.G. Caminsky, E. Mucaki and P.K. Rogan
Reversing differences in chromatin accessibility that distinguish homologous mitotic metaphase chromosomes. W.A. Khan, P.K. Rogan, J.H.M. Knoll
Automated Dicentric Chromosome Identification by Machine Learning-based Image Processing. P.K. Rogan, Y. Li, A. Subasinghe, J. Samarabandu, R. Wilkins and J.H. Knoll
Towards the minimal breast cancer genome and its relevance to chemotherapy. S.N. Dorman, J.H. Knoll, K. Baranova, C. Viner, P.K. Rogan
The FANCM c.5791C>T nonsense mutation (rs144567652) induces exon skipping and is a risk factor for familial breast cancer. Paolo Peterlongo , Francesca Damiola, Eliseos Mucaki, Valentina Dall’Olio ,Sara Pizzamiglio , Irene Catucci , Anders Kvist , Paolo Verderio, Mara Colombo , Loris Bernard , Hans Ehrencrona, Laura Caleca, Valeria Pensotti , Sylvie Mazoyer, Peter K. Rogan ,Paolo Radice
Please contact us if you would like to meet or discuss this work.
September 19. Abstract on metaphase epigenetics: platform presentation at American Society of Human Genetics meeting
Non-random, locus-specific differences in DNA accessibility are present in homologous metaphase chromosomes. W. A. Khan1,3, P. K. Rogan2,3,4, J. H. M. Knoll1,3,4 1) Department of Pathology; 2) Departments of Biochemistry and Computer Science; 3) University of Western Ontario, London, Ontario, Canada; 4) Cytognomix, London, Ontario, Canada.
/
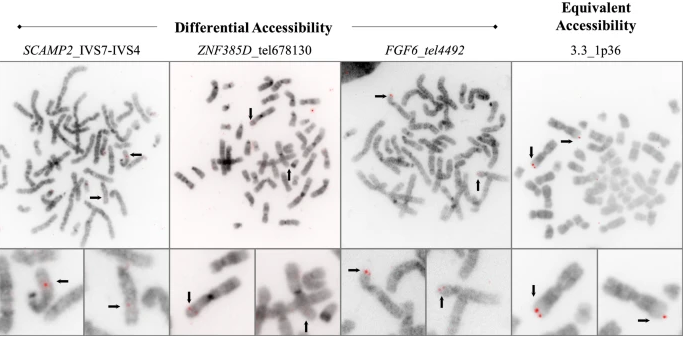
Condensation differences between heterochromatin and euchromatin along the lengths of homologous, mitotic metaphase chromosomes are well known. This study describes differences in metaphase compaction between homologous euchromatic loci. We report molecular cytogenetic data showing local differences in condensation between homologs that are related to differences in accessibility (DA) of associated DNA probe targets. Reproducible DA was observed at ~10% of 450 distinct genomic regions mapped by single copy fluorescence in situ hybridization (scFISH). Fourteen short (1.5-5kb) sc and low copy (lc) FISH probes (from chromosomes 1, 5, 9, 11, 15, 16, 17, 22) targeting genic and non-genic regions with and without DA were developed and hybridized to cells from 10 individuals with cytogenetically-distinguishable homologs. Differences in hybridization were non-random for 6 genomic regions (RGS7, CACNAB1, HERC2, PMP22:IVS3, ADORA2B:IVS1, ACR) and were significantly-biased towards the same homolog (p< 0.01; n = 355 cells). The imprinted paternal chromosome 15 in a three-generation pedigree also showed non-random bias in DA. DNA probes within CCNB1, C9orf66, ADORA2B:Ex 1-IVS1, PMP22:IVS4-Ex 5, and a nongenic region within 1p36.3 did not show DA, while OPCML showed unbiased DA. A subset of probes was mapped onto chromosome topography by FISH-correlated atomic force microscopy (AFM). To quantify DA and pinpoint probe locations, we performed 3D-structured illumination super-resolution microscopy (3D-SIM). 3D anaglyph videos showed genomic regions with DA having nearly 5-fold larger differences in volumetric integrated probe intensities between homologs. Additional non-DA probes (NOMO1, NOMO3) hybridized to grooves in chromosome topography and exhibited a narrow range of probe depths (average: 0.08 μm) along axial and lateral axes of the 2 homologs. In contrast, probe for targets with DA (HERC2, PMP22:IVS3, ACR) significantly differed in probe depth (average: 0.77 μm) and volume (p < 0.05) between each homolog. Interestingly, genomic regions without DA are enriched in epigenetic marks (DHS, H3K27Ac, H3K4me1) of accessible interphase chromatin to a greater extent than regions with DA, suggesting these differences may be correlated with epigenetic marks established during the previous interphase. In summary, we present several lines of evidence that regional differences in condensation between homologs are programmed during metaphase chromosome compaction.
Click here for presentation details: session, location, time.
Customized scFISH probes
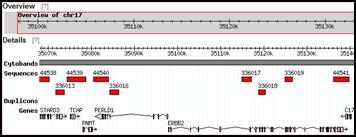
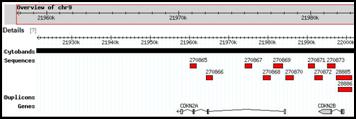 Snapshots of predesigned SC-FISH probes from selected regions of chromosomes 9 and 17
Snapshots of predesigned SC-FISH probes from selected regions of chromosomes 9 and 17
Customized single copy FISH probes can detect small rearrangements in chromosomes with precision, without compromising sensitivity or specificity. Useful for chromosome breakpoint delineation or confirmation of subtle rearrangements suggested by other techniques.
Features
Probes pre-designed for ~223,000 genomic intervals (average genomic resolution ~15 kb)
Available labeled or unlabeled
No Cot-1 or blocking DNA required
Probes detect 100 fold smaller genomic targets than conventional FISH probes. Sizes range from 1.5 to 10 kb
Often multiple probes per gene available
Probes detect rearrangements in small genes
Probe validation by nested PCR or other rapid hybridization methods. Optional FISH validation available.
All probes have been validated by FISH
Research Use Only
Covered by US Pat. No 7,734,424, 8,209,129, and 8,407,013
MutationForecaster
A subscription to our MutationForecaster system provides genome browser access to our genome-wide custom probe designs through the cytogenetic Visual Analytics Decision Support Tool. This tool displays the locations of all of our custom FISH products. Or, contact us for the probes you are looking for.
Automated Dicentric Chromosome Identifier and Dose Estimator
ADCI is available for Windows operating systems (with optional GPU acceleration) or through the internet via Amazon Webservices. See our wiki (link).