We have published a new version of:
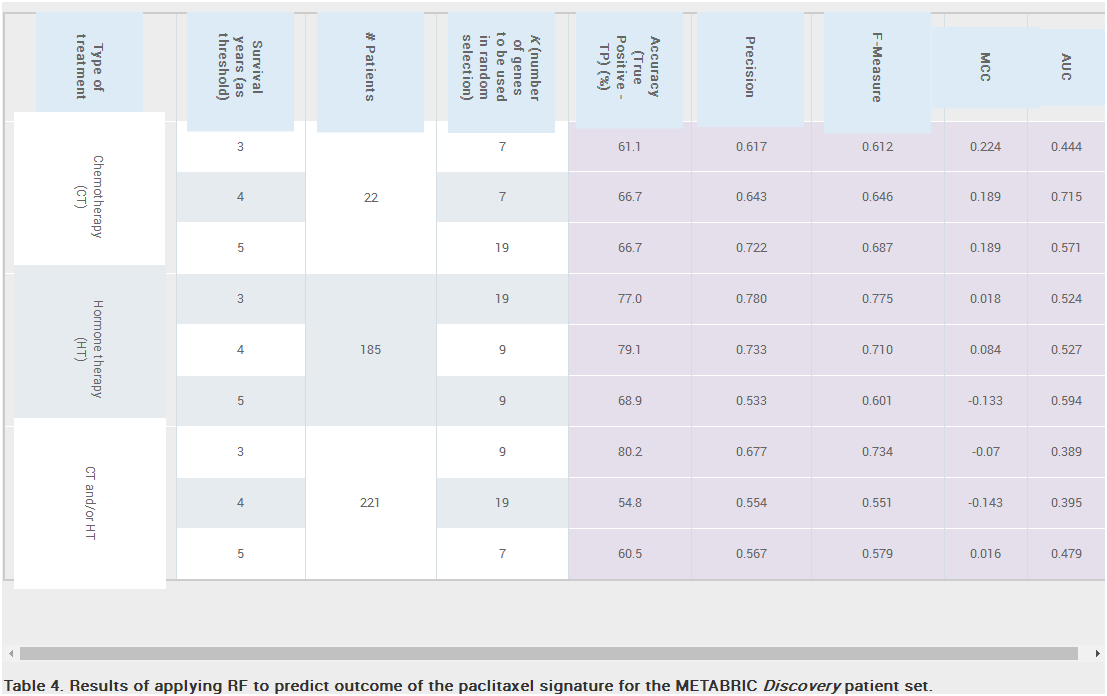
Predicting Outcomes of Hormone and Chemotherapy in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) Study by Biochemically-inspired Machine Learning. F1000Research 2017, 5:2124 (doi:10.12688/f1000research.9417.2)
The revision addresses the comments of the reviewers and adds several new analyses and results. Among our findings was the discovery of significant batch effects that, respectively, differentiate gene expression of signature genes in the Discovery and Validation patient datasets. This is an important cautionary message that should be considered when analyzing the performance of any machine learning based method.