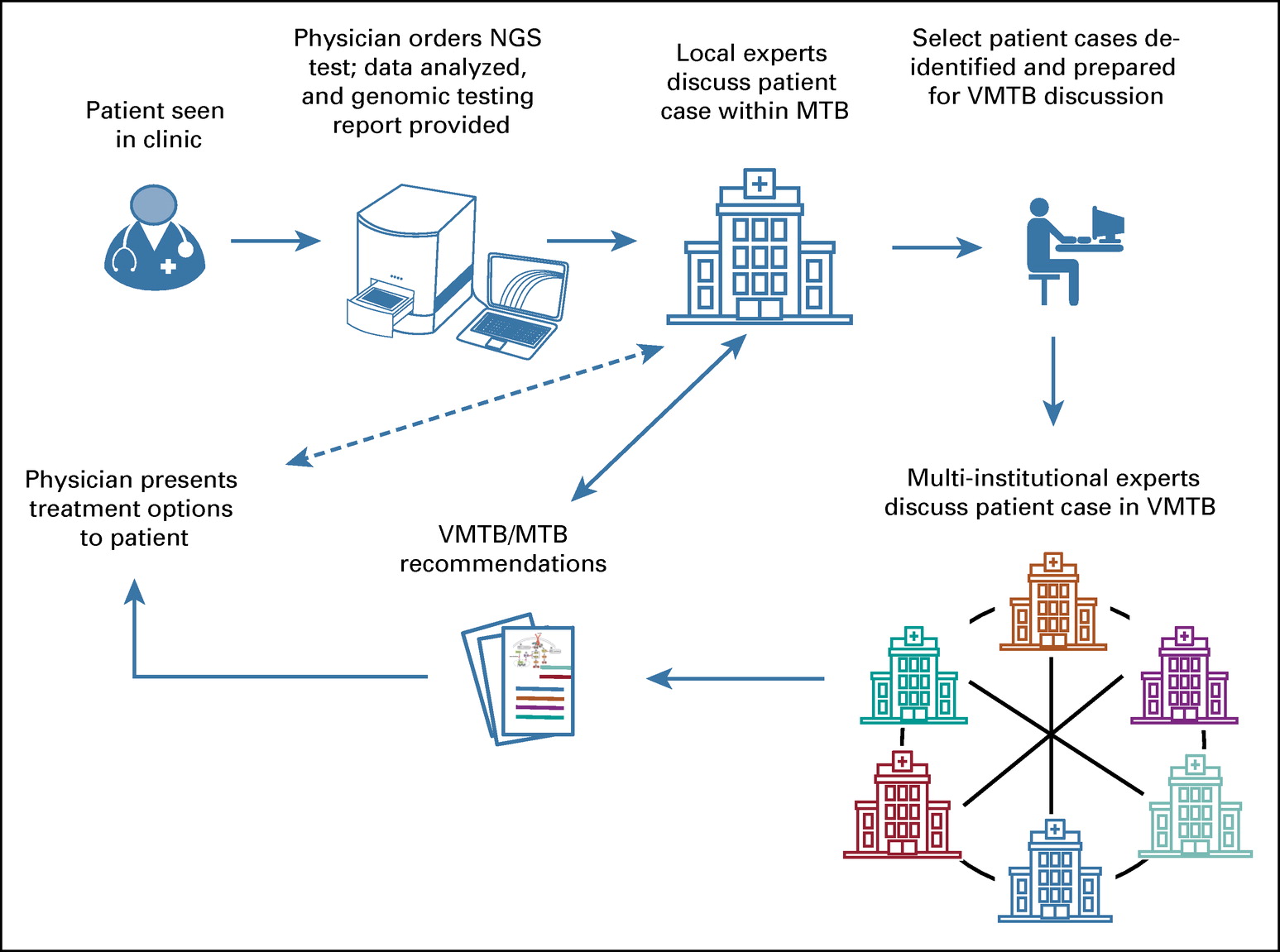
We will be presenting:
Determination of radiation exposure levels by fully automated
dicentric chromosome analysis: Results from IAEA MEDBIODOSE
(CRP E35010) interlaboratory comparison
at both the 19th International Congress of Radiation Research (Aug. 25-29, 2019) and the 12th International Symposium on Chromosome Aberrations (Aug. 27, 2019) in Manchester, UK. This study compared the performance of our Automated Dicentric Chromosome Identifier and Dose Estimator (ADCI) using data from 6 different laboratories. Each of these members of the IAEA-sponsored Cooperative Research Project E35010, submitted images for calibration curve construction and at least 2 samples of unknown exposure to CytoGnomix for analysis with ADCI. We will report the results of this analysis during this presentation.
This poster presentation is now available on the Zenodo website (http://doi.org/10.5281/zenodo.4012749)
doi: DOI 10.5281/zenodo.4012748
Authors:
Rogan P , Shirley B , Li Y , Guogyte K , Sevriukova O , Ngoc Duy P , Moquet J ,
Ainsbury E , Sudprasert W , Wilkins R , Norton F , Knoll J
Department of Biochemistry , University of Western Ontario, London Ontario, Canada
Department of Pathology and Laboratory Medicine, University of Western Ontario, London
Ontario, Canada
Radiation Protection Centre, Ministry of Health (L T -RPC), Vilnius, Lithuania
Dalat Nuclear Research Institute (VN-DNRI), Dalat, Vietnam
Public Health England (PHE), Oxford, Great Britain
Thai Biodosimetry Network, Kasetsart University (THA), Bangkok, Thailand
Health Canada, Ottawa Ontario, Canada
Canadian Nuclear Laboratories, Chalk River Ontario, Canada
Cytognomix, London Ontario, Canada