August 8-12, 2016. Presentation about metaphase chromosome epigenetics
Dr. Peter K. Rogan presented “Reversing chromatin accessibility differences that distinguish homologous mitotic meetaphase chromosomes,”at the Gordon Research Conference on DNA Topoisomerases in Biology and Medicine at Sunday River in Newry, ME, United States.
July 29, 2016. The MutationForecaster Value Proposition
MutationForecaster is catching on. Researchers, clinicians and commercial laboratories are realizing the value of being able to detect and interpret mutations that other platforms miss. Cytognomix has picked up multiple new subscribers from Germany, Switzerland, Australia, China, and Canada this year, and subscription renewals from last year. Cytognomix continues to push the envelope, for the first time publishing papers describing a Unified framework for analyzing gene variants in non-coding and coding gene regions and applying this framework in a large clinical study of inherited breast and ovarian cancer. These reports have led to invitations to contribute our unique expertise to interpretation of results of large inherited cancer genetic studies in the United States and in France. These ongoing projects are showing that the effects of mutations we predict by information theory-based approaches can be confirmed with corresponding gene expression studies in collaborators’ laboratories. What are we working on next for the MutationForecaster suite?
- Adding to our Interactive Report generator to summarize key findings (currently available at MutationForecaster).
- Incorporating our Unified Analytical Framework for complete gene and genome sequence analysis.
- Bespoke Consulting Services to assist you with variant analysis using our software products
This will give our customers will have access to our latest for analysis, filter and interpret their own data. Wouldn’t you like access to these capabilities? Subscribe! NGS sequencing itself may be more accessible and economical today than it has ever been. What we’ve learned from our complete gene sequencing projects is that this success comes with rapidly expanding collections of gene variants, many of which have never been reported before or have been found only rarely. Comprehensive sequencing significantly magnifies the challenges of accurate genome interpretation. Our approach allows you to focus these large collections on only the most functionally relevant variants for review, experimental validation, and prioritization. See what others think of MutationForecaster to gain access to our patented technologies. They are only available from Cytognomix.
July 13, 2016. New publication on Automated Estimation of Radiation Exposure
The paper can be accessed at: abstract or full text (web), or downloadable pdf .
Please contact us if you would like a copy for non-commercial use.
July 1, 2016. Article published on Centromere Detection in Human Metaphase Chromosomes
Our previous preprint in bioRxiv on centromere detection has been published in F1000Research:
Subasinghe A, Samarabandu J, Li Y et al. Centromere detection of human metaphase chromosome images using a candidate based method. F1000Research 2016, 5
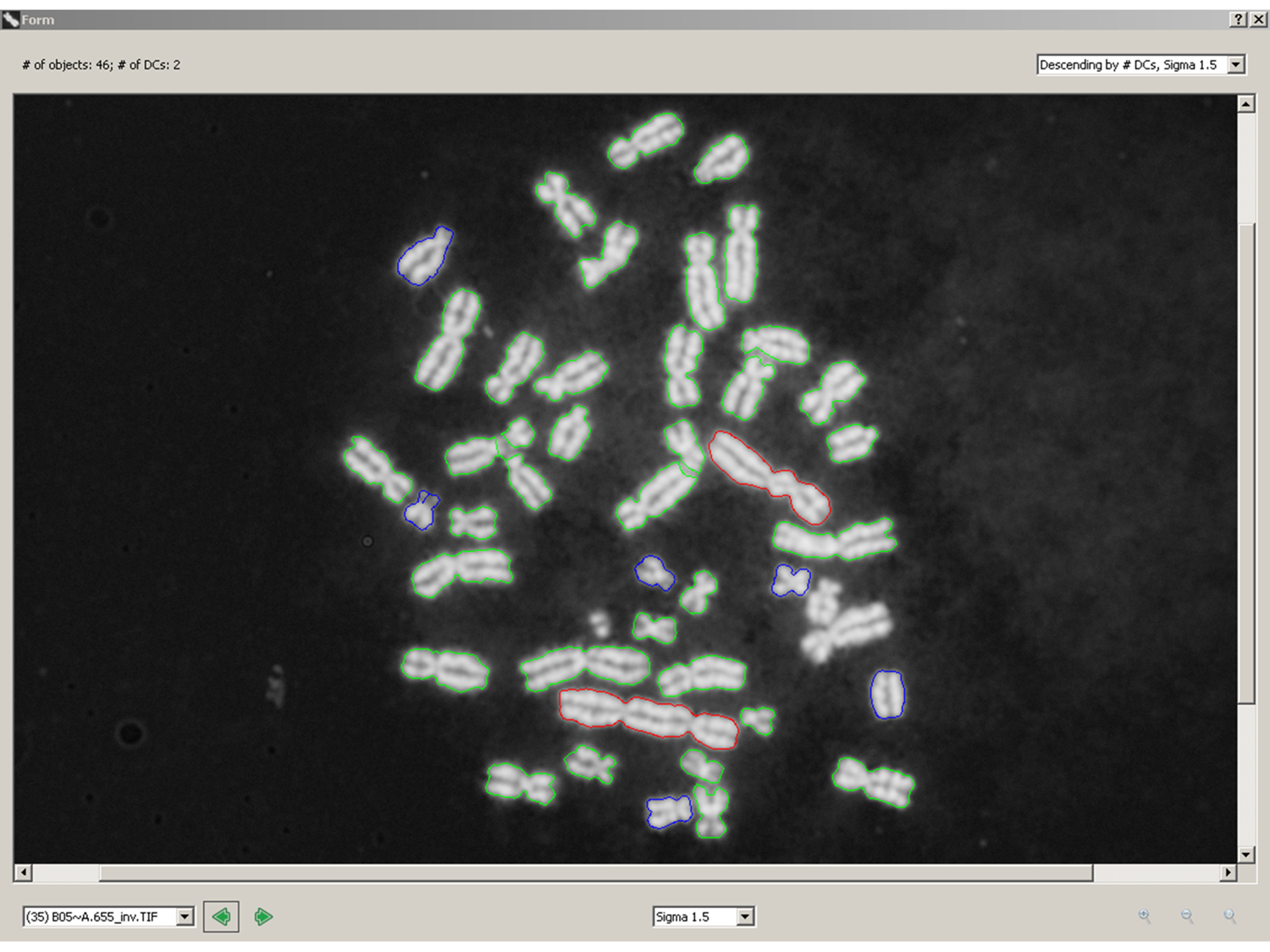
June 6, 2016. New paper on automated biodosimetry to be published
Our paper, “Radiation Dose Estimation by Automated Cytogenetic Biodosimetry” by Peter K. Rogan, Yanxin Li, Ruth Wilkins, Farrah N. Flegal, and Joan H. M. Knoll, has been accepted for publication in the journal Radiation Protection Dosimetry.
Figure 1. Representative processed metaphase image in ADCI:
May 6, 2016. Upcoming public presentations
- Peter K. Rogan, Yanxin Li, Ruth Wilkins, Farrah Flegal, Joan HM Knoll. Radiation Dose Estimation by Automated Cytogenetic Biodosimetry, Great Lakes/Canadian Bioinformatics Conference (CCBC/GLBIO). May 16, 2016. University of Toronto (Platform Presentation).
- Peter K. Rogan. Radiation Dose Estimation by Automated Cytogenetic Biodosimetry. Platform presentation. Great Lakes Chromosome Conference. May 20, 2016. University of Toronto.
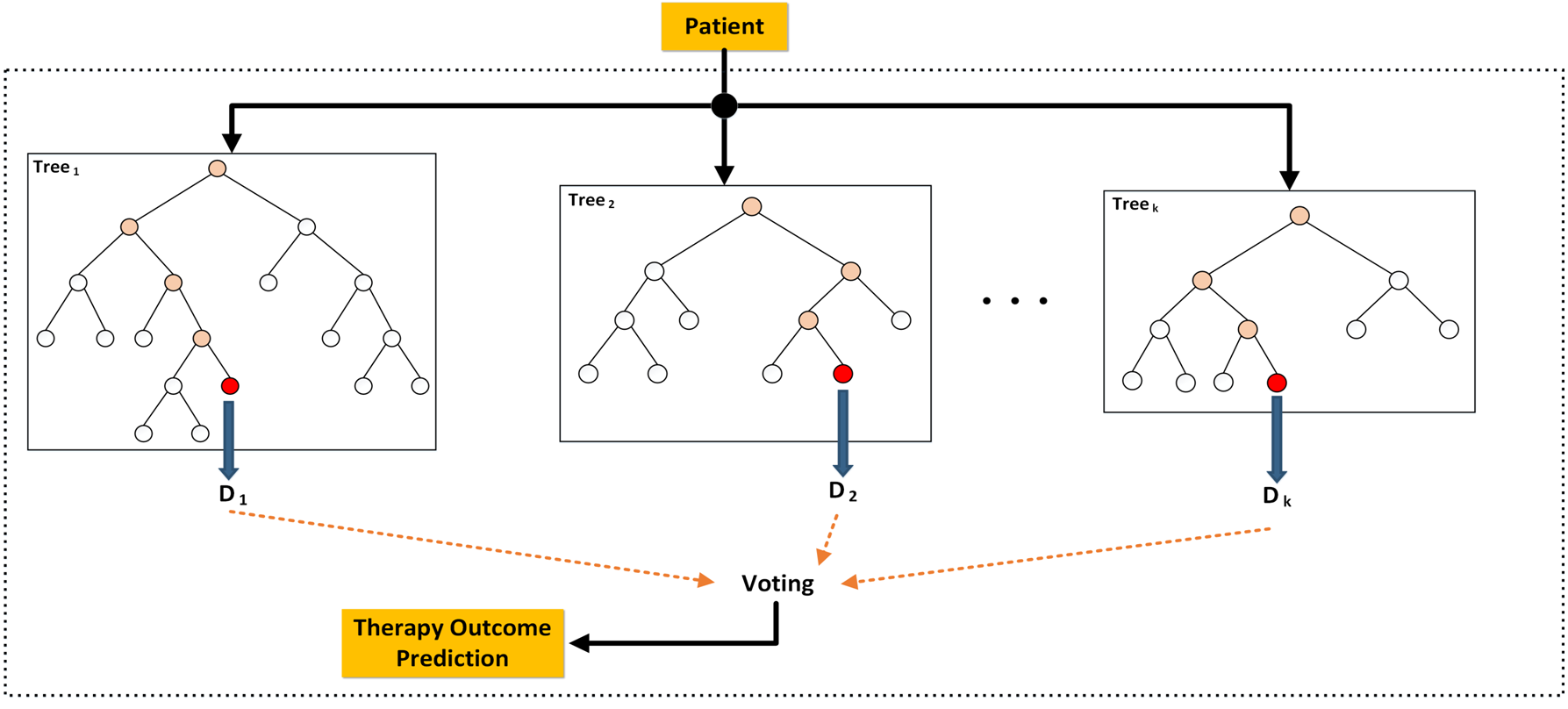
- Peter K. Rogan. Cisplatin Response Prediction in Recurrent Bladder Cancer using Biochemically-inspired Machine Learning. Oral and Poster presentations. 3rd International Molecular Pathological Epidemiology Meeting. May 13, 2016. Dana-Farber Cancer Institute, Boston.
- Rezaeian I, Mucaki E, Baranova K, Quang HP, Angelov D, Ilie L, Ngom A, Rueda L, Rogan PK. Predicting outcome of hormone and chemotherapy in the METABRIC breast cancer study. Great Lakes/Canadian Bioinformatics Conference (GLBIO/CCBC). May 16, 2016. University of Toronto.
- Baranova K, Mucaki EJ, Angelov D, Lizotte D, and Rogan PK. Cisplatin Response Prediction in Recurrent Bladder Cancer using Biochemically-inspired Machine Learning. Great Lakes/Canadian Bioinformatics Conference (GLBIO/CCBC). May 16, 2016. University of Toronto.
- Lu R and Rogan PK. Predicting cis-regulation in human promoters by information density-based clustering of heterotypic transcription factor binding sites. Great Lakes/Canadian Bioinformatics Conference (GLBIO/CCBC). May 16, 2016. University of Toronto.
April 30, 2015. Interview about BMC Medical Genomics and Human Mutation breast cancer articles
Video interview with Eliseos John Mucaki and Peter K Rogan by Cara Campbell on CTV News-London Ontario. April 29, 2016.
(http://london.ctvnews.ca/video?clipId=857860&binId=1.1137789&playlistPageNum=1)
April 28, 2016. Press release
The University of Western Ontario has issued a press release about our studies describing a unified framework for prioritization of mutations in breast and ovarian cancer. 
Other press outlets:
- EurekAlert
- Medical Xpress
- Science Newsline
- Science Codex
- NewsUnited
- MyInforms
- My-News-Site
- BioPortfolio
- Wn.com
- Bioengineer.org
- Ovarian cancer and us
- HealthyTips
- SistersNetwork
- Sci24h
- Spjnews
April 26, 2016. Grant funding received for chromosome structure studies using Cytognomix ab initio scDNA probes
Discovery Grant from Natural Sciences and Engineering Research Council received for “Unravelling the metaphase epigenome: Differences in DNA accessibility between homologous chromosomes.” Joan Knoll, Principal Applicant and Peter Rogan, Co-applicant have been awarded 5 years of funding (2016-21) for this project.
April 11, 2016. New paper on analysis of variants of uncertain significance in hereditary breast & ovarian cancer

Our paper, which describes a generalized information theory-based approach for mutation analysis of protein-nucleic binding sites, has been published:
Mucaki, E, Caminsky N, Perri A, Lu R, Laederach A, Halvorsen, M, Knoll, JHM, Rogan PK. A unified analytic framework for prioritization of non-coding variants of uncertain significance in heritable breast and ovarian cancer, BMC Medical Genomics, 9:19, 2016. DOI: 10.1186/s12920-016-0178-5. (link to paper) (PubMed citation)
March 29, 2016. New publication on cost effectiveness of gene expression microarray testing in cancer diagnosis
Through a pan-Canadian collaboration led by Greg Zaric, we have published:
Cost-effectiveness of using a gene expression profiling test to aid in identifying the primary tumour in patients with cancer of unknown primary. M B Hannouf, E Winquist, S M Mahmud, M Brackstone, S Sarma, G Rodrigues, P Rogan, J S Hoch and G S Zaric.
The Pharmacogenomics Journal advance online publication 29 March 2016; doi: 10.1038/tpj.2015.94 (Link)
March 15, 2015. Funding from the Build-in-Canada Innovation Program
Cytognomix pre-qualifies for funding from Build in Canada Innovation Program. Our proposal to test the Automated Dicentric Chromosome Identifier with Health Canada and Canadian Nuclear Laboratories has been approved by Public Works and Government Services Canada (link).
March 10, 2016. New paper accepted on prioritization strategy for gene variants of uncertain significance in breast/ovarian cancer
Our paper:
“A unified analytic framework for prioritization of non-coding variants of uncertain significance in heritable breast and ovarian cancer,” by
Eliseos J. Mucaki; Natasha G. Caminsky; Ami M. Perri; Ruipeng Lu; Alain Laederach; Matthew Halvorsen; Joan H.M. Knoll; and Peter K. Rogan
has been accepted for publication in the journal, BMC Medical Genomics.
A preprint of this article is currently available at BioRxiv: http://biorxiv.org/content/early/2015/11/11/031419.
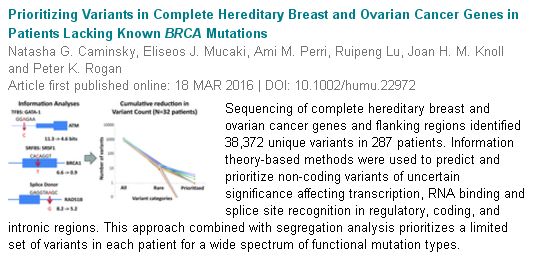
February 16, 2016. New publication on inherited breast and ovarian cancer
Our new paper on interpretation of gene variants in inherited breast and ovarian cancer has been accepted for publication in the journal, Human Mutation as a Research Article.
“Prioritizing variants in complete Hereditary Breast and Ovarian Cancer (HBOC) genes in patients lacking known BRCA mutations,” by Natasha G. Caminsky1, Eliseos J. Mucaki1, Ami M. Perri1, Ruipeng Lu2, Joan HM. Knoll3,4 and Peter K. Rogan1,2,4,5.
1Department of Biochemistry, Schulich School of Medicine and Dentistry, Western University, London, Canada, N6A 2C1, 2Department of Computer Science, Faculty of Science, Western University, London, Canada, N6A 2C1, 3Department of Pathology and Laboratory Medicine, Schulich School of Medicine and Dentistry, Western University, London, Canada, N6A 2C1, 4Cytognomix Inc. London, Canada, 5Department of Oncology, Schulich School of Medicine and Dentistry, Western University, London, Canada, N6A 2C1
A preprint of this article is published at http://biorxiv.org/content/early/2016/02/09/039206
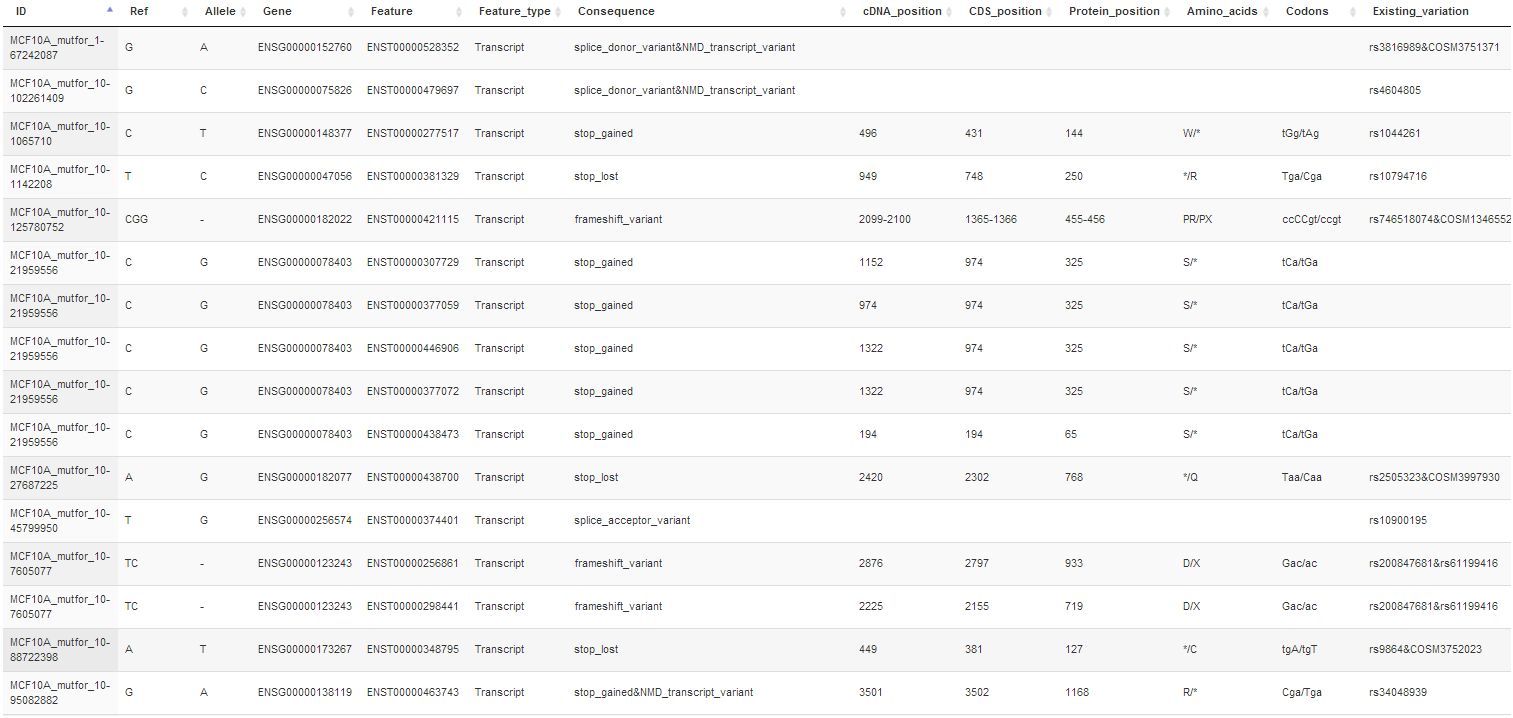
Feb. 15, 2016. Improved filtering in Mutation Forecaster for Variant Effect Predictor
We have added new capabilities to Variant Effect Predictor. Exome sequencing reveals many variants that have little or no effect on phenotype. You can remove these variants in MutationForecaster with our new stringency filters. Different default levels of filtering are offered. These can also be customized based on allele frequencies, predicted SIFT, Polyphen, variant type (eg. synonymous change), or protein coding domain containing the variant.